The spectrum of an object can give us a lot of information, such as type of the object, radial velocity, redshift, presence of chemical elements or compounds in its atmosphere, an estimate of its distance, etc. In this article we take a look at one-dimensional spectra created by SDSS and saved in FITS files.
I suppose the reader has at least some basic knowledge about spectroscopy, redshift, SDSS and FITS files. If this is not the case, you can refer to:
Inspecting the spectrum in a FITS file
I have already explained in another post, three different methods to download the spectrum of an object from SDSS servers. By creating an instance of the SpecObj class from the sdss python package and passing the specObjID, you can download the spectrum as a FITS file by calling the download_spec method:
import sdss sp = sdss.SpecObj(320932083365079040) sp.download_spec()
Now, we have a file named spec-0285-51930-0184.fits in our working directory. A good option to handle FITS files is using astropy.io.fits package. Let’s import the neccessary libraries and open the downloaded file.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy import units as u
hdul = fits.open('spec-0285-51930-0184.fits')
hdul.info()
Filename: spec-0285-51930-0184.fits No. Name Ver Type Cards Dimensions Format 0 PRIMARY 1 PrimaryHDU 142 () 1 COADD 1 BinTableHDU 26 3839R x 8C [E, E, E, J, J, E, E, E] 2 SPECOBJ 1 BinTableHDU 262 1R x 126C [6A, 4A, 16A, 23A, 16A, 8A, E, E, E, J, E, E, J, B, B, B, B, B, B, J, 22A, 19A, 19A, 22A, 19A, I, 3A, 3A, 1A, J, D, D, D, E, E, 19A, 8A, J, J, J, J, K, K, J, J, J, J, J, J, K, K, K, K, I, J, J, J, J, 5J, D, D, 6A, 21A, E, E, E, J, E, 24A, 10J, J, 10E, E, E, E, E, E, E, J, E, E, E, J, E, 5E, E, 10E, 10E, 10E, 5E, 5E, 5E, 5E, 5E, J, J, E, E, E, E, E, E, 25A, 21A, 10A, E, E, E, E, E, E, E, E, J, E, E, J, 1A, 1A, E, E, J, J, 1A, 5E, 5E] 3 SPZLINE 1 BinTableHDU 48 29R x 19C [J, J, J, 13A, D, E, E, E, E, E, E, E, E, E, E, J, J, E, E]
The fits.open function returns an HDUList object. We used the info method of this object to view the summarized content of our file. As you see in the above results, the FITS file consists of a primary HDU (hdul[0]) and three extensions (hdul[1], hdul[2], hdul[3]). The primary HDU contains only the Header keywords. The three extensions are of type BinTableHDU, each of them with its own header and data.
To see the column names of each table, use the columns attribute. Wavelength and flux of the spectrum are stored in the first extension (hdul[1]). Let’s see all columns of this extension:
cols1 = hdul[1].columns print(cols1)
ColDefs(
name = 'flux'; format = 'E'
name = 'loglam'; format = 'E'
name = 'ivar'; format = 'E'
name = 'and_mask'; format = 'J'
name = 'or_mask'; format = 'J'
name = 'wdisp'; format = 'E'
name = 'sky'; format = 'E'
name = 'model'; format = 'E'
)
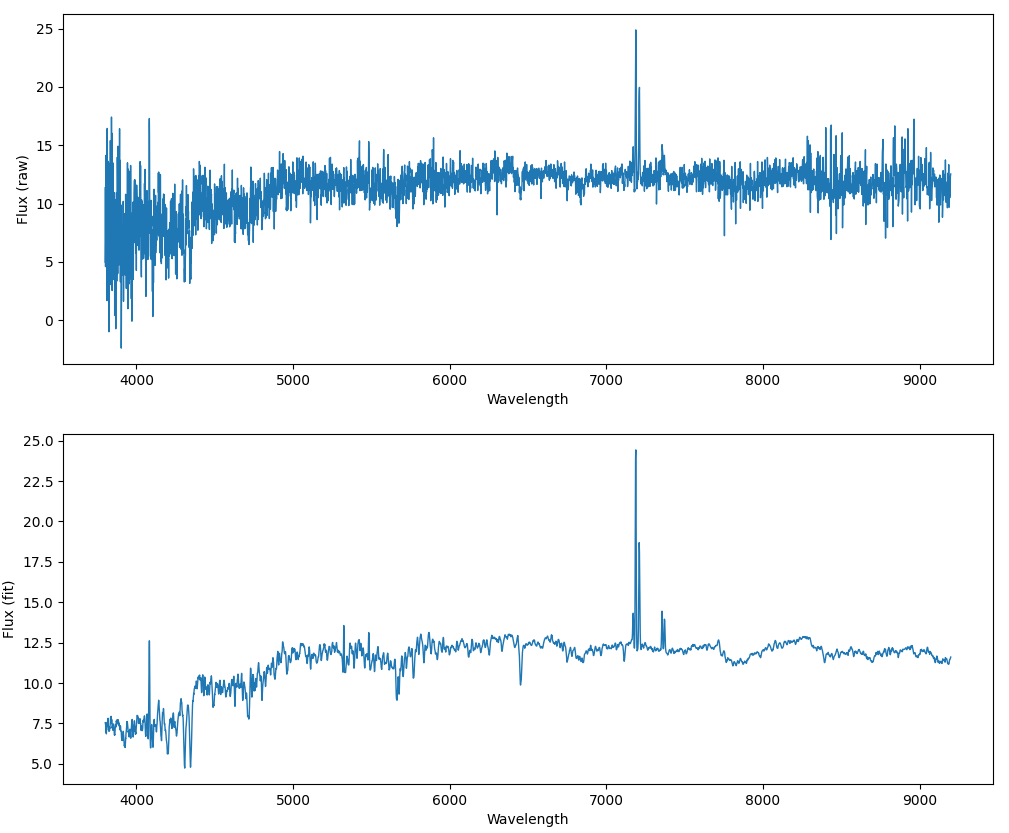
The wavelengths are stored in the loglam column in the logarithmic scale. The flux column is the raw flux data as recorded by the spectroscope, and the model column is the result of the best fitted model on the flux. In most cases we prefer to use model instead of flux.
To access the data stored in each extension, we should first use the data attribute. Then we can access values of each columns by using field method and passing the column name. Let’s extract wavelength, flux and model columns:
data1 = hdul[1].data
wavelength = 10 ** data1.field('loglam')
flux = data1.field('flux')
model = data1.field('model')
print('Wavelength :', wavelength)
print('Flux (raw) :', flux)
print('Model (fit):', model)
Wavelength : [3801.0188 3801.8933 3802.77 ... 9193.905 9196.0205 9198.141 ] Flux (raw) : [ 4.972567 11.401632 4.585463 ... 10.810025 12.317144 12.532938] Model (fit): [ 7.54909 7.5063486 7.429691 ... 11.473688 11.527207 11.608632 ]
It is a good practice to use appropiate units when working with scientific data. The SDSS measures wavelength in Angstrom (AA) and the unit of flux in:
![]()
So, let’s recreate these three variables with their appropriate units:
wavelength = wavelength * u.Unit('AA')
flux = flux * 10**-17 * u.Unit('erg cm-2 s-1 AA-1')
model = model * 10**-17 * u.Unit('erg cm-2 s-1 AA-1')
Now, we plot both flux and model on y-axis and wavelength on x-axis.
fig, ax = plt.subplots(2, 1, figsize=(12,10))
ax[0].plot(wavelength, flux, linewidth=1)
ax[1].plot(wavelength, model, linewidth=1)
ax[0].set_xlabel('Wavelength')
ax[1].set_xlabel('Wavelength')
ax[0].set_ylabel('Flux (raw)')
ax[1].set_ylabel('Flux (fit)')
plt.show()
The emission lines
Up to here, we have just worked with the first extension of the FITS file. The second extension just gives us some general information, such as coordinates, class of object, redshift, etc. Most of these could be requested by SQL scripts directly from the SDSS servers, so we don’t explain them here.
The third extension is important for our project. This is where the information about emission lines can be found. In fact, the plot of a spectrum without indication of spectral lines doesn’t give us much information. Let’s see column names of the third extension:
cols3 = hdul[3].columns data3 = hdul[3].data print(cols3)
ColDefs(
name = 'PLATE'; format = 'J'
name = 'MJD'; format = 'J'
name = 'FIBERID'; format = 'J'
name = 'LINENAME'; format = '13A'
name = 'LINEWAVE'; format = 'D'
name = 'LINEZ'; format = 'E'
name = 'LINEZ_ERR'; format = 'E'
name = 'LINESIGMA'; format = 'E'
name = 'LINESIGMA_ERR'; format = 'E'
name = 'LINEAREA'; format = 'E'
name = 'LINEAREA_ERR'; format = 'E'
name = 'LINEEW'; format = 'E'
name = 'LINEEW_ERR'; format = 'E'
name = 'LINECONTLEVEL'; format = 'E'
name = 'LINECONTLEVEL_ERR'; format = 'E'
name = 'LINENPIXLEFT'; format = 'J'
name = 'LINENPIXRIGHT'; format = 'J'
name = 'LINEDOF'; format = 'E'
name = 'LINECHI2'; format = 'E'
)
Here, we are specially interested in four of these columns: LINENAME, LINEWAVE, LINEZ and LINEAREA. These are the names of the lines, the rest wavelengths of the lines, the calculated redshift from each line, and areas of the lines, respectively. Let’s extract these columns in four new variables and create a pandas DataFrame from them:
line_names = data3.field('LINENAME')
line_waves = data3.field('LINEWAVE')
line_z = data3.field('LINEZ')
line_area = data3.field('LINEAREA')
df = pd.DataFrame(
{'name': [i for i in line_names],
'lam_rest': [i for i in line_waves],
'z': [i for i in line_z],
'area': [i for i in line_area]
}
)
df = df[abs(df['area'])>0]
print(df)
name lam_rest z area 6 [O_II] 3725 3727.091727 0.094871 30.996830 7 [O_II] 3727 3729.875448 0.094871 24.527514 8 [Ne_III] 3868 3869.856797 0.094871 -3.518241 9 H_epsilon 3890.151080 0.094871 -8.486563 10 [Ne_III] 3970 3971.123187 0.094871 -22.393383 11 H_delta 4102.891631 0.094871 -6.722156 12 H_gamma 4341.684313 0.094871 3.061384 13 [O_III] 4363 4364.435300 0.094871 1.015044 14 He_II 4685 4686.991444 0.094871 -1.985071 15 H_beta 4862.682994 0.094871 9.303711 16 [O_III] 4959 4960.294901 0.094870 6.017811 17 [O_III] 5007 5008.239638 0.094871 18.227947 18 He_II 5411 5413.024423 0.094871 -2.460578 19 [O_I] 5577 5578.887704 0.094872 -1.581697 20 [O_I] 6300 6302.046377 0.094872 10.753190 21 [S_III] 6312 6313.805533 0.094870 0.442675 22 [O_I] 6363 6365.535420 0.094871 4.292485 23 [N_II] 6548 6549.858929 0.094871 15.137597 24 H_alpha 6564.613894 0.094870 83.739006 25 [N_II] 6583 6585.268445 0.094870 54.291546 26 [S_II] 6716 6718.294208 0.094871 23.901695 27 [S_II] 6730 6732.678076 0.094871 12.859660 28 [Ar_III] 7135 7137.757103 0.094871 -1.571996
We have deleted the rows whose line areas were zero because we don’t need them. Note that the wavelengths of the lines are rest or emitted wavelengths. These are the values that we already know from the laboratories on the Earth. But in the spectrum we should draw the observed wavelength of lines. According to the definition of the redshift:
![]()
So,
![]()
Using the above formula, we can calculate the observed wavelength of the lines easily:
df['lam_obs'] = df['lam_rest'] * (1 + df['z'])
Now we are ready to create the plot of the spectrum.
fig, ax = plt.subplots(figsize=(12,8))
label_y = np.random.uniform(
low=model.min().value*1.1,
high=model.max().value*0.9,
size=len(df)
)
ax.plot(wavelength, model, linewidth=1)
for i in range(len(df)):
ax.axvline(x=df['lam_obs'].iloc[i],
color='r',
alpha=0.3,
label=df['name'].iloc[i],
ls='--',
lw=0.7)
ax.text(x=df['lam_obs'].iloc[i] ,
y=label_y[i],
s=df['name'].iloc[i],
fontsize='small',
rotation=90)
plt.xlim(4000, 8000)
plt.show()
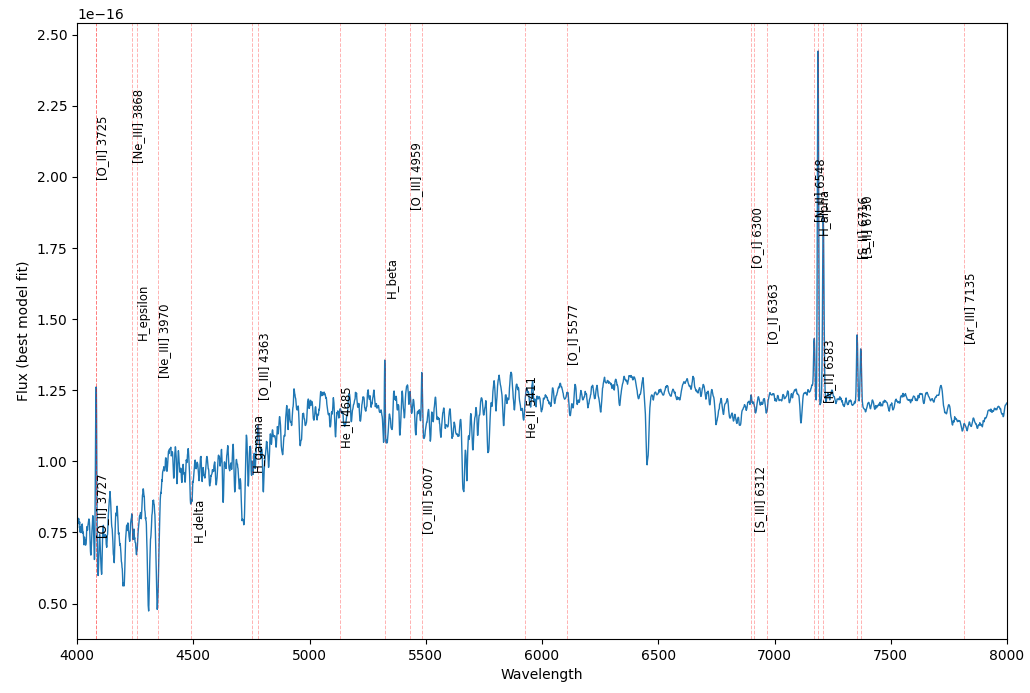
Not bad. But the problem is that when the line are so close to each other they can not be distinguished. This is why we need an interactive plot that can be zoomed in and out.
Making the plot interactive
In order to create an interactive plot we use bokeh python package. There are also other libraries such as plotly that could be used in the same way. First, we should import necessary libraries and create the plot in almost similar way. Here, we can use some features such as hover tools that let’s the user to view some useful information.
from bokeh.plotting import figure, show
from bokeh.models.tools import HoverTool
from bokeh.models import Span, Label
p = figure(title="Spectrum",
sizing_mode="stretch_width",
x_range=(4000, 8000),
tools=[HoverTool(), 'pan', 'wheel_zoom', 'reset'],
tooltips="lambda=@x{0.000} | flux=@y",
x_axis_label="Wavelength", y_axis_label="Flux")
p.line(wavelength, model, line_color="blue", line_width=1)
for i in range(len(df)):
ver = Span(location=df['lam_obs'].iloc[i],
dimension='height',
line_color='red',
line_dash='dashed',
line_width=1)
p.add_layout(ver)
lbl = Label(x=df['lam_obs'].iloc[i],
y=label_y[i],
text=df['name'].iloc[i])
p.add_layout(lbl)
show(p)
You can find the complete source code here.

